
Index
1. Home/Query page
2. Results page
Analysis Parameter Summary
Re-Analysis Box
Tag Clouds
Documents
Biomedical Terms
3. Data Integration and Bioinformatics Analysis
4. Co-occuerence analysis
Contents
1. Home/Query page [Go to top]
The home page of bioTextQuest is the page where the user can input his query: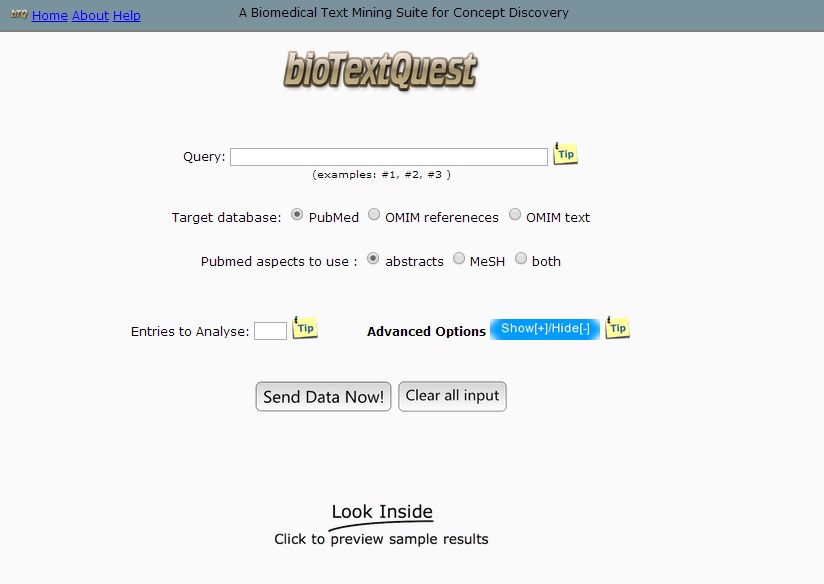
Query: The keywords with which the search will be performed over the OMIM and Medline databases. The query should be in the format that is normally accepted by the database and will be processed exactly as it would be processed if the search was performed from the original web site. Any notation that would be acceptable in the original database interface is acceptable here, e.g. p53 AND (leaver OR pancr*) NOT cancer (Medline).
It should be noted that the abstracts are kept in a local database that is updated every week with new records from Medline and OMIM. Thus, some searches may not yield the same number of articles when performed in the bioTextQuest server and the PubMed web site.
Entries to Analyse: The maximum number of papers that should be analyzed by bioTextQuest. If there are more papers than the limit imposed by the user, bioTextQuest retrieves the more recent ones.
Advanced Options: Control of stemming and clustering algorithms and their parameters.
Upon Clicking on the Advanced Options, the user can change the default options:
Stemming Algorithm: (Default: no stemming) Words of common root may be stemmed if the user chooses so (e.g. 'process', 'processed' and 'processing' may be stemmed to the root 'process'). Currently only the Porter's stemming algorithm is supported.
Clustering Algorithm: (Default: K-Means). Clustering can be performed by various algorithms. Currently the following clustering algorithms are supported: Markov Clustering (MCL), Hierarchical - Average Linkage, K-means, Spectral Clustering, Affinity Propagation and RNSC.
Each Clustering algorithm has parameters whose optimum values (according to the of the author of the implementation) are chosen as default. The user can change them according to his needs using the slider.
- Markov Clustering (MCL), parameter: inflation (default: 1.8)
The rule of thumb is that the larger the inflation parameter the more clusters will be created and the more fine-grained they will be. - Hierarchical - Average Linkage, parameter: number of clusters (default: 3)
The user specifies the desired number of resulting clusters. - K-Means, parameter: number of clusters (default: 3)
The user specifies the desired number of resulting clusters. - Spectral Clustering (MCL), parameter: epsilon (default: 1.03)
The rule of thumb is that the larger the epsilon parameter the more clusters will be created and the more fine-grained they will be.
Clustering can be performed according to various similarity methods such as: Cosine similarity, Tanimoto similarity, Pearson, Kendall and Spearman correlations as well as Okabi BM25 document ranking algorithm.
2. Results page [Go to top]
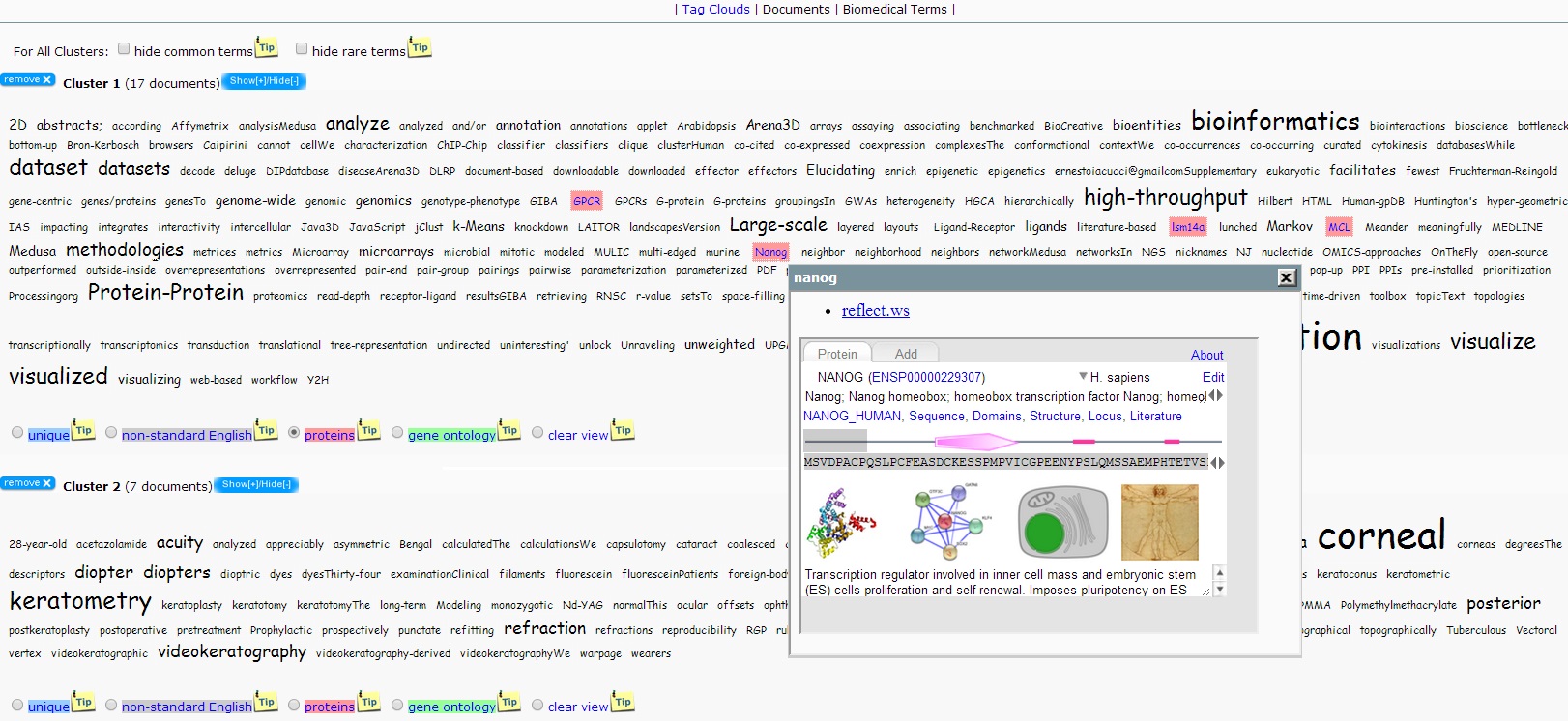
Analysis Summary
The Analysis Summary displays the parameters, limits and keywords used to make the query whose results are being displayed
Export
The links provided in this section enable users to download tab-delimited files containing either the Cluster terms or the PubMed IDs per cluster for further analysis.
New Analysis
The New Analysis Box allows the user to rerun the same query after changing the parameters and excluding any Biomedical terms they wish to exclude.
Tag Clouds
The clusters created by bioTextQuest. Each cluster is represented by keywords. If a cluster is expanded, its keywords are displayed as a tag cloud. The size of each term is proportional to its frequency.
The user may choose to highlight the terms that fall in any of the following 4 categories in a cluster:
- unique
significant terms that do not appear in any other cluster. - non-standard English
significant terms that are not standard grammatical terms (they do not belong in the dictionary used by bioTextQuest). - proteins
significant terms that describe proteins. These terms are annotated using the Reflect tool (http://reflect.ws/) - processes / pathways / cellular components.
significant terms that are also GO (gene ontology) terms. These terms are annotated using EBI's Gene Ontology (http://www.ebi.ac.uk/GO/).
- hide common terms
terms that are common in ALL clusters. This options is hidden when there is only a cluster - hide rare terms
terms that are rare in each cluster (naturally, a term might be rare in a cluster and have a high occurence in another).
The user can remove some of the clusters before proceeding with a new analysis.
Documents

The documents that belong to each cluster.
- Clicking on the
 icon will open a new window in PubMed with the respective article.
icon will open a new window in PubMed with the respective article.
- Clicking on [abstract] will show a popup with the abstract.
- Clicking on [Reflected abstract] will show a popup with the reflected abstract.
Biomedical Terms
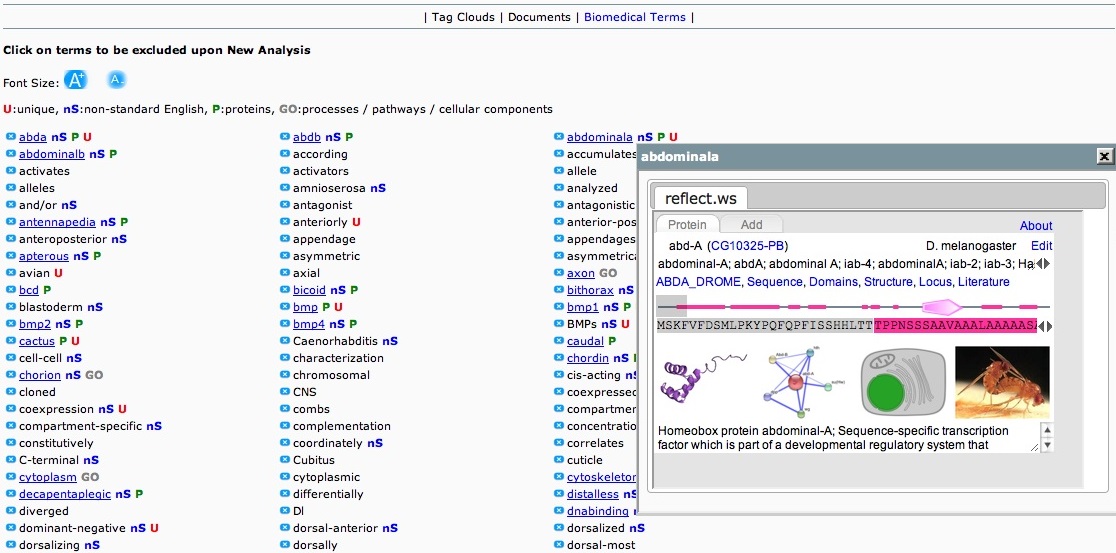
All the terms (go-list Terms) that were used to cluster the documents. Each term is annotated depending on whether it belongs in any of the four categories
The user can remove some of the terms before proceeding with a new-analysis.
3. Data Integration and Bioinformatics Analysis [Go to top]
BioTextQuest offers a variety of bioinformatics analysis and data integration techniques for the tagged biooentities in a selected set of abstracts. To do that we merge BioTextQuest service with the in-house BioCompendium service.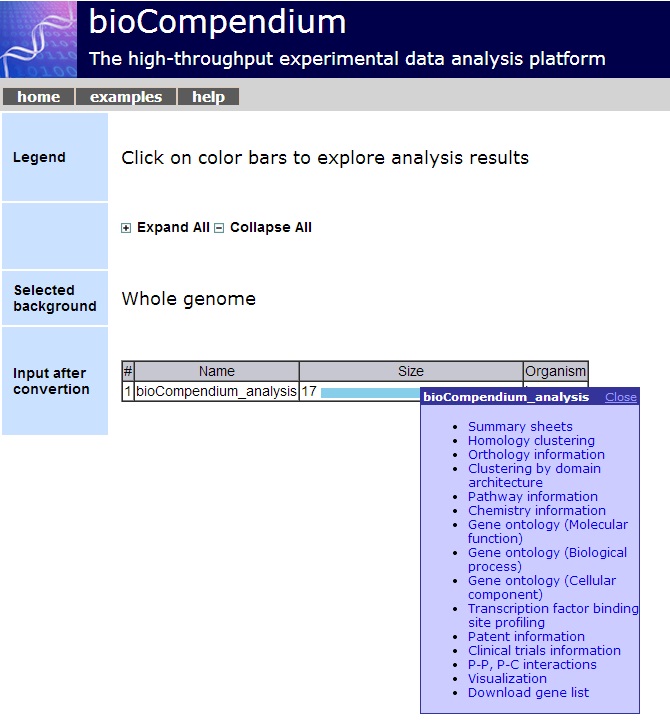
4. Co-occurence Analysis [Go to top]
BioTextQuest offers the ability to select one or more terms and retrieve the abstracts where these terms are co-mentioned. On the Tag Cloud view just right click up to 5 terms which will appear in the new analysis text-box on your left. Alternatively you can manually type the terms of interest in the text box. We distinguish between the abstract-based and sentence-based co-occurrence analysis. One can retrieve the articles and their abstracts where the selected terms are co-mentioned in the abstract. Users can hide or expand the abstracts to see the highlighted terms. In the case of sentence-based co-occurrence analysis users can isolate the abstracts where the terms of interest are co-mentioned in a sentence or even isolate the sentences themselves.